Research
I have been researching in the Clinical Natural Language Processing (NLP) space for the past decade. The overarching theme of my work is semantic parsing of clinical data (like Electronic Medical Records) to extract information that can be used to make actionable decisions for healthcare delivery and contribute to downstream research.
- At Arizona State (2014-20), I worked under Dr. Chitta Baral in the Cognition and Intelligence Lab throughout my BS and MS, where I collaborated with organizations like the Mayo Clinic
- At the University of Illinois (2020-24) as a medical student, I worked under Dr. Yoga Varatharajah on semantic processing of EEG-related medical notes.
- As a resident at Mayo Clinic (2024-), I work in the BEACON Lab on combining my clinical and technical backgrounds to develop, evaluate, and deploy clinical AI tools.
Articles
- Combining interest in artificial intelligence with medicine - a feature in Mayo Clinic about my goals throughout residency and thoughts about the future of AI in medicine
- Some slides from a talk I gave a few months ago on how I use LLMs (for research, clinical practice, and programming).
- Carle Illinois Machine Learning System for EEG Analysis Wins IEEE Honors - Work focusing on extracting components from a clinical specification from epilepsy medical records; won "Best Paper" award at IEEE SPMB 2021.
- Improving AI Models for Better Patient Care - Work presented at NeurIPS 2022
Software
🌵 In addition to papers, I build Clinical AI software. My goals are to
- Make information deep within medical records more accessible: semantically filter, search, and extract clinical information from medical data like Electronic Medical Records
- Is generalizable: can be applied across a range of clinical tasks without significant task-specific training
- Develop packages, models, and tools that can be used as building blocks for further downstream clinical research and development
software:
- * Osler: a workspace + copilot for physicians writing H&P notes that is designed to augment thinking, not replace it. (More information)
- MedQA: a GPT-powered clinical reference tool that can answer clinical questions and follow-up questions in natural language, along with references to sources. (More information)
- clinisift: multitool for processing clinical medical records.
- clinitokenizer: sentence tokenizer for clinical/medical text.
models:
- bert-Clinical-NER: a Named Entity Recognition model for clinical entities (problem, treatment, test).
- bert-Med-NER: a Named Entity Recognition model for medication entities (medication name, dosage, duration, frequency, reason).
datasets and resources:
- Clinical GPT-3: exploration of OpenAI GPT-3's abilities to perform various Natural Language Processing tasks in the clinical domain, on patient medical records.
- Sample Medical Notes: a collection of generated medical notes that can be used publicly for demos, etc. Coming soon.
Selected Works
Full work on Google Scholar, my resume, or LinkedIn.
MIRIAD: Augmenting LLMs with millions of medical query-response pairs [preprint]
Q. Zheng, S. Abdullah, S. Rawal, C. Zakka, S. Ostmeier, M. Purk, E. Reis, E. J. Topol, J. Leskovec, M. Moor. arXiv (cs.CL), 2025.
MIRIAD is a large-scale curated corpus of ~5.8M medical instruction-response pairs grounded in peer-reviewed medical literature, designed to improve retrieval-augmented generation (RAG) and reduce hallucinations in medical QA. We also introduce MIRIAD-Atlas, an interactive semantic map to explore queries and grounded responses across medical disciplines.
Paper (arXiv) | Project page | Code | Dataset | Demo
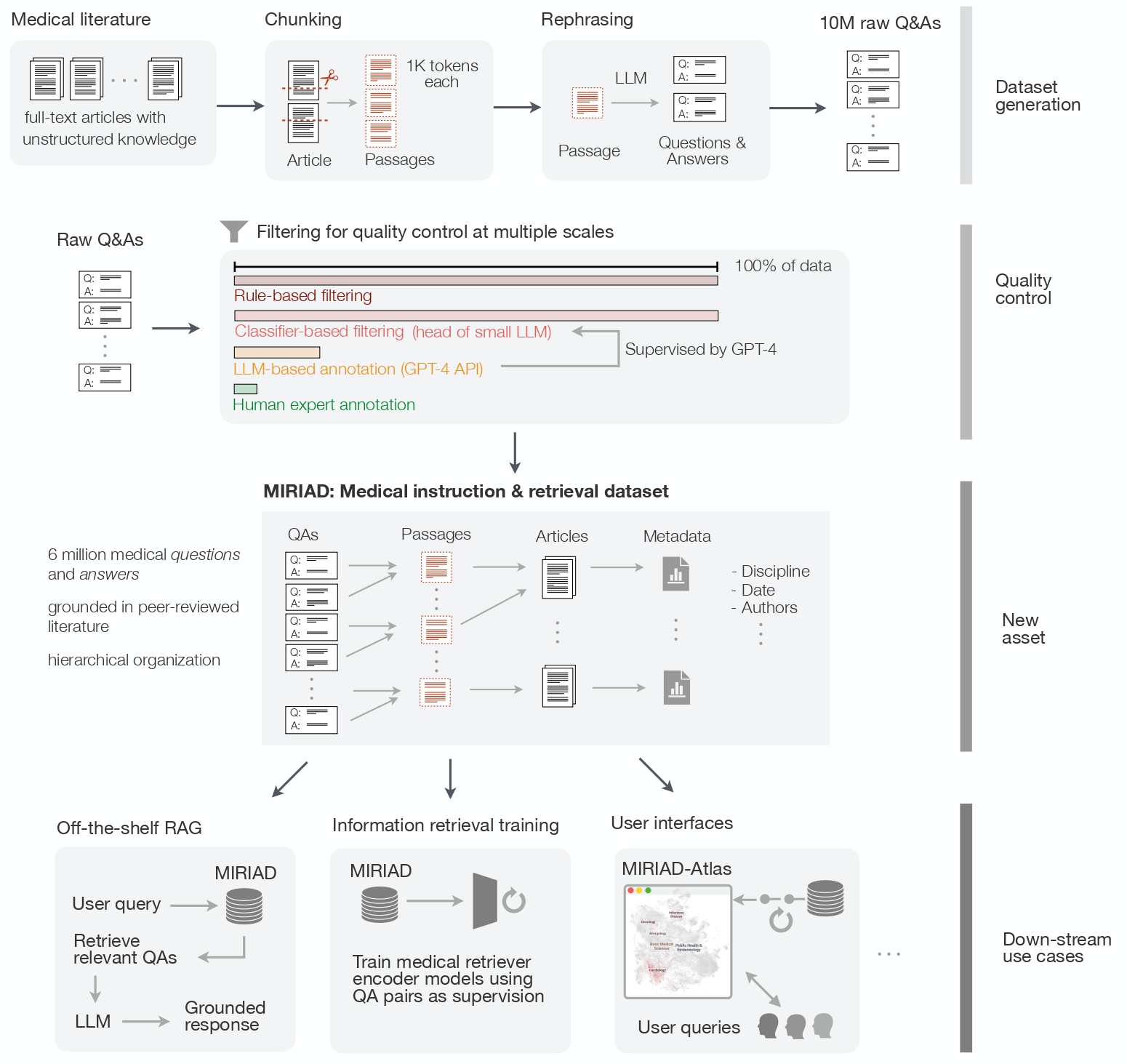
SCORE-IT: A Machine Learning-based Tool for Automatic Standardization of EEG Reports
S Rawal, Y Varatharajah. 2021 IEEE Signal Processing in Medicine and Biology Symposium (SPMB).
In this work, we propose a machine learning-based system that automatically extracts components from the SCORE specification from unstructured, natural-language EEG reports. Specifically, our system identifies (1) the type of seizure that was observed in the recording, per physician impression; (2) whether the session recording was normal or abnormal according to physician impression; (3) whether the patient was diagnosed with epilepsy or not.
🏆 Best Paper Award at 2021 IEEE Signal Processing in Medicine and Biology Symposium (SPMB).
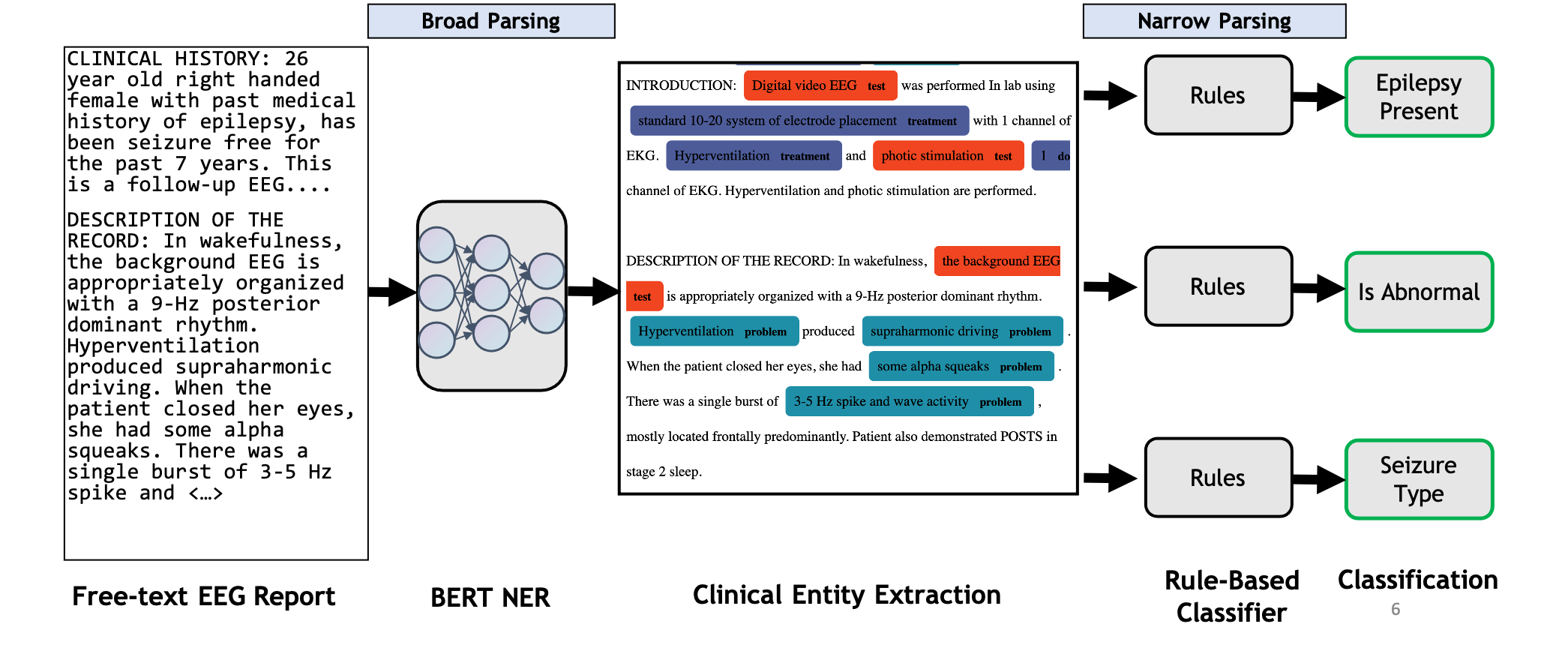
Evaluating Latent Space Robustness and Uncertainty of EEG-ML Models under Realistic Distribution Shifts
N. Wagh, J. Wei, S. Rawal, B. M. Berry, Y. Varatharajah. Conference on Neural Information Processing Systems (NeurIPS) 2022.
Abstract: The recent availability of large datasets in bio-medicine has inspired the development of representation learning methods for multiple healthcare applications. Despite advances in predictive performance, the clinical utility of such methods is limited when exposed to real-world data. This study develops model diagnostic measures to detect potential pitfalls before deployment without assuming access to external data. Specifically, we focus on modeling realistic data shifts in electrophysiological signals (EEGs) via data transforms and extend the conventional task-based evaluations with analyses of a) the model's latent space and b) predictive uncertainty under these transforms. We conduct experiments on multiple EEG feature encoders and two clinically relevant downstream tasks using publicly available large-scale clinical EEGs. Within this experimental setting, our results suggest that measures of latent space integrity and model uncertainty under the proposed data shifts may help anticipate performance degradation during deployment.
PDF on OpenReview via NeurIPS 2022
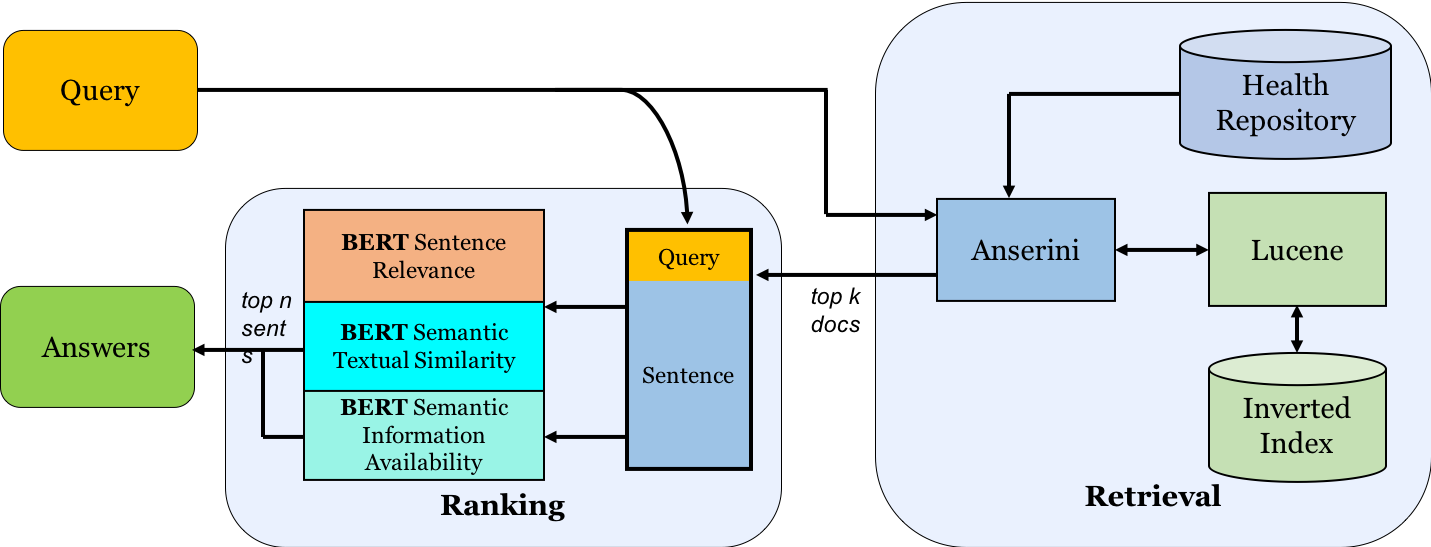
Multi-Perspective Biomedical Semantic Question-Answering (MS Thesis)
This work introduces the concept of a Multi-Perspective IR system, a novel methodology that combines multiple Transformers-based deep learning and traditional IR models to better predict the relevance of a query-sentence pair, along with a standardized framework for tuning this system.
Given a query in natural language, search across 29 million PubMed abstracts and identify top n candidate sentences that answer the query. To better "understand" and rank candidates, a weighted "Multi-Perspective" approach, utilizing three BERT models trained on different tasks, is taken.
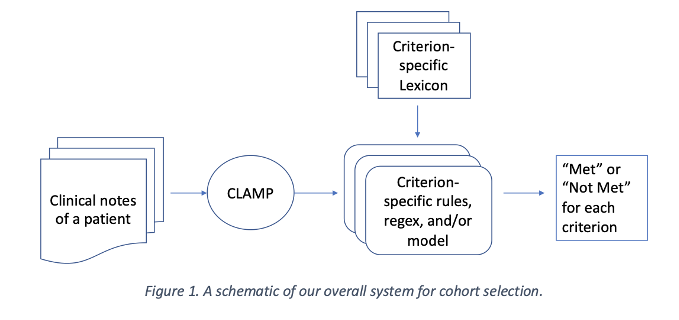
Developing and Using Special-Purpose Lexicons for Cohort Selection from Clinical Notes
S. Rawal, A. Prakash, S. Adhya, S. Kulkarni, S. Anwar, C. Baral, M. Devarakonda. 2018 National NLP Clinical Challenges shared tasks.
Selecting cohorts for a clinical trial requires costly and time-consuming manual chart reviews resulting in poor participation. From natural-language patient medical records, our system classifies whether a patient is within or outside 13 clinical trial cohorts (i.e. alcohol abuse, drug abuse, MI within past 6 months, advanced coronary artery disease).
Part of the n2c2 2018 Challenge – ranked #1 out of 47 teams.
Prescription Information Extraction from Electronic Health Records (BS Thesis)
Bidirectional LSTM + CRF neural architecture for Named Entity Recognition applied to the i2b2 2009 Medication Information extraction challenge.
